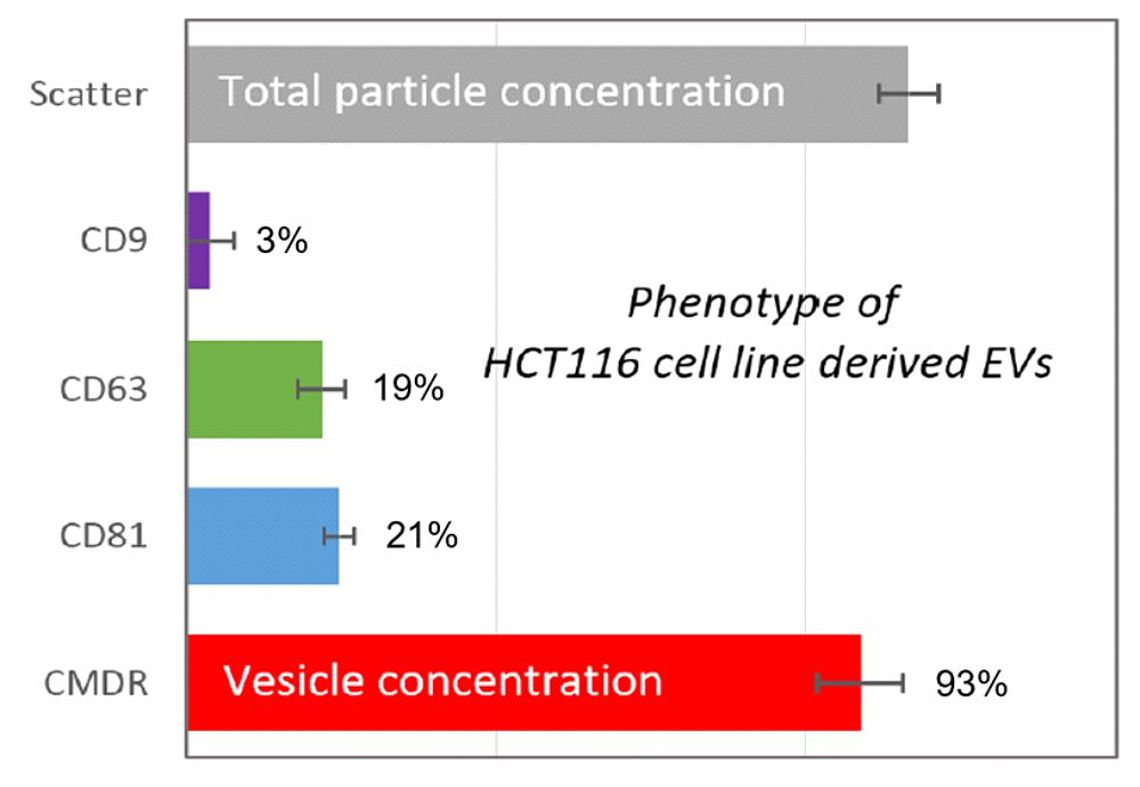
Figure 1: Examination of an extracellular vesicle preparation from the colon cancer cell line HTC116. Comparison between the total particle count (grey; 100%), CD9 positives (violet; 3%), CD63 positives (green; 19%), CD81 positives (blue; 21%) and the biological vesicle count (red; 93%).
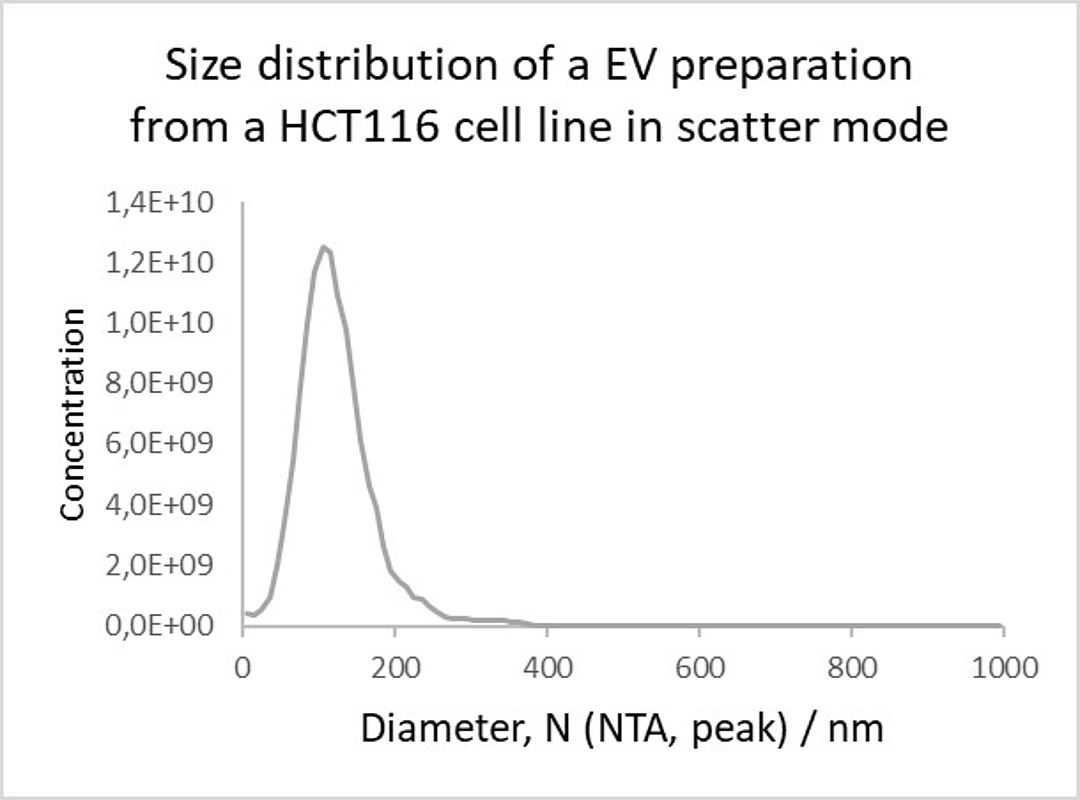
Figure 2: Size distribution of an EV preparation from an HCT116 cell line in scatter mode.
Abstract
Nanoparticle Tracking Analysis (NTA) has emerged as the state-of the-art method for size and concentration characterization of exosomes and extracellular vesicles (Konoshenko et al., 2018, Giebel & Helmbrecht, 2017, Soo et al., 2012). In combination with fluorescence detection (f-NTA) the technology now enables the user to take a deeper look inside dedicated exosome populations and subpopulations (Desgeorges et al., 2020, Rahbari et al., 2019, Weber et al., 2019). Here we demonstrate a quick and easy method for exosome identification by antibody mediated, fluorescence-based nanoparticle tracking analysis in four fluorescent channels with the Particle Metrix ZetaView® instrument.
Introduction
Exosomes are small sized (30 to 150 nm), membrane surrounded extracellular vesicles (EVs) which are ubiquitously secreted by biological cells (Raposo et al., 2013, Yanez-Mo et al., 2015) and present in all kingdoms of life. Stanly and colleagues discovered them in plants (Stanly et al., 2016) whereas Deatherage & Cookson found their presence in bacterial systems (Deatherage & Cookson, 2012). Exosomes play an important role in many cellular processes like maintaining homeostasis (Baixauli et al., 2014), cell-to-cell com-munication (Mathivanan et al., 2010), the metastasis of tumors (Becker et al., 2016), inflammatory processes (Console et al., 2019) and a lot of other pathophysiological activities (Record et al., 2014). Due to their involvement in different important cellular functions, extracellular vesicles and exosomes are currently being investigated in many laboratories all over the world.
Tetraspanins are a vast protein superfamily with multiple members in humans and other species such as Drosophila melanogaster and Caenorhabditis elegans (Huang et al., 2005). They consist of a very flexible extracellular domain, a transmembrane region with four helices and a highly conserved cytoplasmic domain (Andreu and Yáñez-Mó, 2014). Beside their presence in various types of endocytic membranes including the plasma membrane (Berditchevski & Odintsova, 2007) they are highly enriched in extracellular vesicles and exosomes where they built so called tetraspanin-enriched microdomains (TEMs) (Andreu and Yáñez-Mó, 2014). In recent years CD9, CD37, CD63, CD81 and CD82 were primarily used as exosomal markers but it should be noted that exosomes are a very heterogeneous group of vesicles which can differ drastically in their tetraspanin composition (Andreu and Yáñez-Mó, 2014).
Here we report a new, fast and reliable method to specifically identify and differentiate extracellular vesicles and exosomes based on their tetraspanin content (CD9, CD63 and CD81) by using the Particle Metrix ZetaView® QUATT (Meerbusch, Germany). The unique scanning technology of the instrument and the simultaneous use of four high-power lasers (λ1 = 405nm; λ2 = 488nm; λ3 = 520nm; λ4 = 640nm) offers unrevealed sensitivity and a maximum flexibility in fluorescence detection in up to four channels.
Methods
Lyophilized exosomes derived from the colon cancer cell line HTC116 (Rajput et al., 2008; HansaBioMed, Estonia) have been resuspended with ultra-pure water (Carl Roth, Germany) at 1µg/µl as described in the manufacturer’s protocol. The unspecific, lipophilic membrane dye CellMask™ Deep Red (CMDR, Thermo Fisher, USA) was prediluted to a final concentration of 5 µg/ml, also in ultra-pure water (Carl Roth, Germany). Fluorescently labelled antibody conjugates directed against human CD9 (with Alexa Fluor® 405) and human CD81 (with Alexa Fluor® 488) were purchased from R&D Systems (Minneapolis, USA) and used in a final concentration of 0.02 mg/ml. Anti-human CD63-DyLight550 conjugate were obtained from Novus Biologicals (Centennial, USA) and used at 0.088 mg/ml. 6 µg of re-suspended EVs were thoroughly mixed with 1 µl pre-diluted CMDR, 1 µl anti-CD9-Alexa Fluor®405, 1 µl CD63-DyLight550 and 1 µl of CD81-Alexa Fluor® 488 and incubated for 2 hours at room temperature in the dark. The staining mix was then diluted 1:1.000 with practically particle free 1 X PBS (Thermo Fisher, USA) to accomplish a total vesicle concentration of about 5.0 x 107 particles/ml. To evaluate the total particle count and the overall size the stained sample was measured in scatter mode using the 488 nm laser and standard instrument settings (sensitivity: 80, shutter: 100, min. brightness: 30; min. area: 10; max. area: 1000). After that, the specific measurements for anti-CD9-Alexa Fluor® 405 (405 nm violet laser), anti-CD81-Alexa Fluor® 488 (488 nm blue laser), Anti- CD63-DyLight550 (520 nm green laser) and CMDR (640 nm red laser) were carried out using the Particle Metrix ZetaView QUATT® in fluorescence mode with low-bleach technology and standard fluorescence settings (sensitivity: 92, shutter: 100, min. brightness: 25; min. area: 10; max. area: 1000). The resulting videos were analysed with the ZetaView® software 8.05.10 (Particle Metrix, Germany).
The purity was calculated according to the following relationship:
conc. fluorescence mode CMDR
purity = ————————————
conc. scatter mode
The ratios of CD9, CD63 and CD81 positive extracellular vesicles were calculated with the following formulas:
conc. fluorescence 405 nm
% CD9 positives = ———————————— * 100%
conc. scatter mode
conc. fluorescence 488 nm
% CD81 positives = ———————————— * 100%
conc. scatter mode
conc. fluorescence 520 nm
% CD63 positives = ———————————— * 100%
conc. scatter mode
Results & Conclusions
Extracellular vesicle preparations from most origins (e.g. blood, urine etc.) are composed of different vesicles and the vesicles originate from different cells types (Vlassov et al., 2012). Unfortunately, the well-established scatter based NTA technology is not able to discriminate between the different components of those preparations (Desgeorges et al., 2020; Zhang et al., 2019) As described in Methods, we labelled a commercially available EV sample derived from the colon cancer cell line HTC116 simultaneously with the lipophilic membrane stain CMDR and the three tetraspanin antibodies against CD9, CD63 and CD81, respectively. The concentration in scatter mode reflects 100 % of the present particles (see grey bar in figure 1). The data derived from CellMask™ Deep Red demonstrate that the commercial EV sample has a high purity of about 93% (red bar in Figure 1). This is in good agreement with data we achieved with other commercially available extracellular vesicle preparations obtainable from the same supplier (data not shown). However, the tetraspanins were found in lower concentrations. CD9 could only be detected in 3% of the overall particles (violet bar, Figure 1). CD63 and CD81 are present in 19% and 21% of the total vesicles (blue and green bar in Figure 1). Figure 1: Examination of an extracellular vesicle preparation from the colon cancer cell line HTC116. Comparison between the total particle count (grey; 100%), CD9 positives (violet; 3%), CD63 positives (green; 19%), CD81 positives (blue; 21%) and the biological vesicle count (red; 93%).
Simultaneously performed size measurements showed that the membranous vesicles have a slightly smaller mean diameter (95,5 nm) than the total particles (113,9 nm). A very likely explanation for this discrepancy is that bigger non-vesicular impurities such as nanobubbles, inorganic salt precipitates or protein aggregates shift the size distribution and therefore also the mean diameter to a bigger size. However, 80% of the measured particles showed sizes between 60 and 170 nm (see Figure 2). This is in good agreement with previous published results from Ren and colleagues which measured the total particle sizes of HCT 116 cell line derived vesicles between 60 and 150 nm (Ren et al., 2019). Nevertheless, the exosomalmarker positive vesicles showed significantly smaller mean diameter (CD9 = 60 nm, CD63 =78,4 nm and CD81 = 64,6 nm) as already shown for other exosomes by different techniques (Bachurski et al., 2019). Figure 2: Size distribution of an EV preparation from an HCT116 cell line in scatter mode.
Taken together, these results show that simultaneous labelling with specific monoclonal antibodies can identify, discriminate and phenotype exosomes from other extracellular vesicles and non-vesicular nanoparticles. Additionally, the concurrent staining with four markers leads to a more precise result as the sample-to-sample variation is eliminated compared with single stainings. Last but not least this new technology saves time and sample.
References
Andreu Z. and Yáñez-Mó M. Tetraspanins in Extracellular Vesicle Formation and Function. Front Immunol. 2014; 5: 442
Bachurski D., Schuldner M., Nguyen P-H., Malz A., Reiners K.S., Grenzi P.C., Babatz F., Schauss A.C., Hansen H.P., Hallek M. and Pogge von Strandmann E. Extracellular vesicle measurements with nanoparticle tracking analysis – An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell Vesicles. 2019: 8(1): 1596016.
Baixauli F., Lopez-Otin C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014; 5:403.
Becker A., Thakur B., Weiss J. M., Kim H. S., Peinado H., Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016 Dec 12;30(6):836-848.
Berditchevski F. and Odintsova E. Tetraspanins as Regulators of Protein Trafficking. Traffic 2007; 8:89–96.
Console L., Scalise M., Indiveri C. Exosomes in inflammation and role as biomarkers. Clin Chim Acta. 2019; 488:165-171.
Deatherage B.L. and Cookson B.T. Membrane Vesicle Release in Bacteria, Eukaryotes, and Archaea: a Conserved yet Underappreciated Aspect of Microbial Life. Infect Immun. 2012; 80(6):1948-57.
Desgeorges A., Hollerweger J., Lassacher T., Rohde E., Helmbrecht C., and Gimona M. Differential fluorescence nanoparticle tracking analysis for enumeration of the extracellular vesicle content in mixed particulate solutions. Methods. 2020 Feb 17. pii: S1046-2023 (19)30264-6.
Giebel B. and Helmbrecht C. Methods to Analyze EVs; Methods Mol Biol. 2017; 1545:1-20.
Huang S., Yuan S., Dong M., Su J., Yu C., Shen Y., Xie X., Yu Y., Yu X., Chen S., Zhang S., Pontarotti P. and Xu A. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics. 2005 Dec; 86(6):674-84.
Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int. 2018: 8545347.
Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteom. 2010;73(10):1907–1920.
Rahbari M, Pecqueux M, Aust D, Stephan H., Tiebel O. Chatzigeorgiou A., Tonn T., Baenke F., Rao V., Ziegler N., Greif H., Lin K., Weitz J., Rahbari N.N. and Kahlert C. Expression of Glypican 3 is an Independent Prognostic Biomarker in Primary Gastro-Esophageal Adenocarcinoma and Corresponding Serum Exosomes. J Clin Med. 2019;8(5):696.
Rajput A, Dominguez San Martin I, Rose R, Beko A., Levea C., Sharratt E., Mazurchuk R., Hoffman R.M., Brattain M.G. and Wang J. Characterization of HCT116 human colon cancer cells in an orthotopic model. J Surg Res. 2008;147(2): 276–281.
Raposo G. and Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383.
Record M., Carayon K., Poirot M., Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841(1):108–120.
Ren R., Sun H., Ma C. , Liu J. and Wang H. Colon cancer cells secrete exosomes to promote self-proliferation by shortening mitosis duration and activation of STAT3 in a hypoxic environment. Cell Biosci. 2019; 9:62
Soo C.Y., Song Y., Zheng Y., Campbell E.C., Riches A.C., Gunn-Moore F. and Powis S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012, 136:192–197.
Stanly C., Fiume I., Capasso G., Pocsfalvi G. Isolation of Exosome-Like Vesicles from Plants by Ultracentrifugation on Sucrose/Deuterium Oxide (D2O) Density Cushions. Methods Mol Biol. 2016; 1459:259-69.
Vlassov A.V., Magdaleno S., Setterquist R. and Conrad R. Exosomes: Current knowledge of their composition, biological functions, anddiagnostic and therapeutic potentials. Biochimica et Biophysica Acta (BBA). 2012 Jul; 1820(7): 940-948
Weber A., Wehmeyer J.C., Schmidt V., Lichtenberg A. and Akhyari P. Rapid Fluorescence-based Characterization of Single Extracellular Vesicles in Human Blood with Nanoparticle-tracking Analysis. J Vis Exp. 2019 Jan 7;(143)
Yanez-Mo M., Siljander P.R., Andreu Z., Zavec A.B., Borras F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., Colas E., Cordeiro-da Silva A., Fais S., Falcon-Perez J.M., Ghobrial I.M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N.H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Kramer-Albers E.M., Laitinen S., Lasser C., Lener T., Ligeti E., Line A., Lipps G., Llorente A., Lotvall J., Mancek-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-’t Hoen E.N., Nyman T.A., O’Driscoll L., Olivan M., Oliveira C., Pallinger E., Del Portillo H.A., Reventos J., Rigau M., Rohde E., Sammar M., Sanchez-Madrid F., Santarem N., Schallmoser K., Ostenfeld M.S., Stoorvogel W., Stukelj R., Van der Grein S.G., Vasconcelos M.H., Wauben M.H., De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015; 4:27066.
Zhang Z., Tang C., Zhao L., Xu L., Zhou W., Dong Z., Yang Y., Xiec Q. and Fang X. Aptamer-based fluorescence polarization assay for separation-free exosome quantification. Nanoscale, 2019, 11: 10106-10113
Back
